BIO-HELIX - MB202-P040
AmpFAST 2X PCR SuperMix (5-30 sec/kb; ≤6kb, 1X)
Size: 40 rxns
AmpFAST 2X PCR SuperMix was premixed with an optimized concentration of AmpFAST Taq DNA Polymerase, dNTPs, Mg2+, and reaction buffer that could be used in extremely fast PCR experiments as well as massive gene detection requirements, which also performs better amplification functions for GC-rich and complicated secondary structures. For shorter sequence or plasmid samples, it is suggested to use 5 sec/kb elongation speed or less circuits to shorten your PCR reacting time. While for longer sequences (≥3 kb) or complicated samples that usually have lesser PCR products, it is suggested to use 15-30 sec/kb elongation speed or more circuits. Meanwhile, an “A” base at the 3’-end of the PCR products amplified by this enzyme can be directly used in TA cloning.
Save time without sacrificing yield! AmpFAST 2X PCR SuperMix delivers superior performance in fast or high-throughput PCR, producing clear bands even for difficult templates. Ready-to-use convenience, exceptional speed, and TA cloning compatibility make it the ideal choice for modern molecular biology workflows.
✅ Ultra-fast amplification: 5–30 sec/kb for ≤6 kb targets
✅ Pre-mixed 2X formula: add template & primers only
✅ Optimized for GC-rich and complex secondary structures
✅ Generates A-overhangs for direct TA cloning
✅ Perfect for colony PCR, plasmid verification, gene screening
✅ Stable and reproducible performance for massive detection workflows
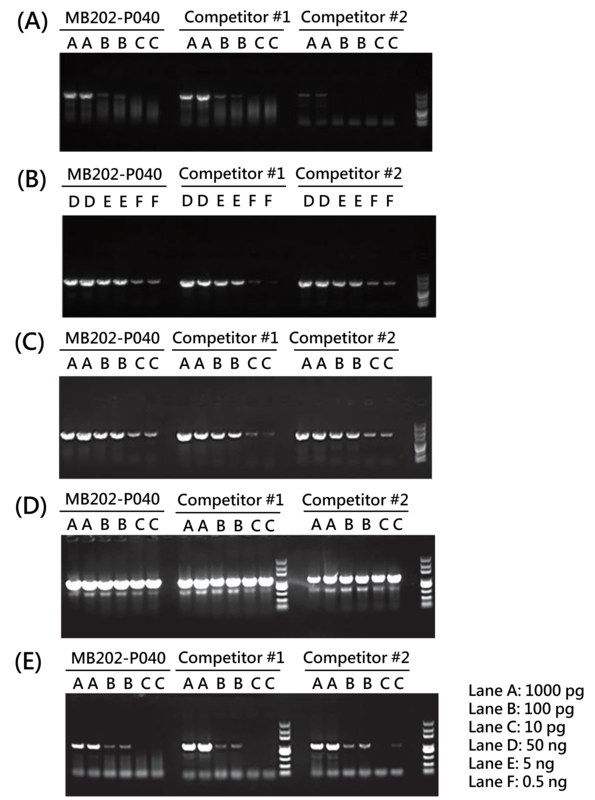
Figure 1. Amplification Results Using MB202-P040 on Templates from Different Sources
(A) E. coli (3 kb) (B) Soybean Genomic DNA (1.5 kb) (C) Mouse Genomic DNA (3 kb) (D) λDNA (1.2 kb) (E) Mouse Genomic DNA (1 kb, GC-rich)
Groups A-C were amplified at a speed of 15 sec/kb, while groups D-E were amplified at 5 sec/kb.
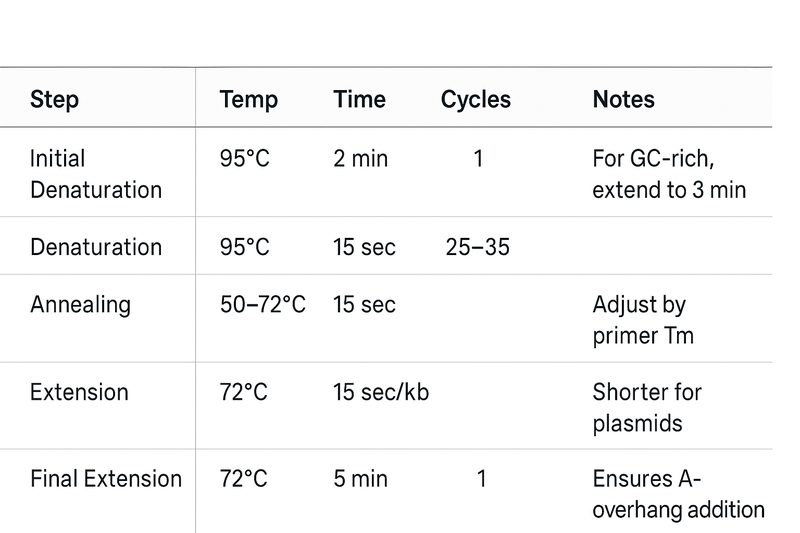
Note: Because PCR efficiency depends on template complexity, primer design, GC-content, and the thermal cycler’s characteristics (ramp-rate, block uniformity), users are advised to optimize annealing temperatures, extension times, and cycle numbers for each new primer–template combination and instrument model.
Tip: If unusual template (e.g., very high GC content, long amplicon, crude lysate) or a different thermal-cycler is used, consider increasing extension time or performing a gradient annealing temperature trial.
| Name | Download |
|---|---|
| PROTOCOL | Bio-Helix_MB202-P040_Protocol_V1.pdf |
| Safety Data Sheet|SDS | Bio-Helix_MB202-P040_MSDS_V1.pdf |